[Tutorial] SCN2A mutations in neurodevelopmental disorders
Tutorial for Programming basics (Chapter 3)
1. Introduction
1.1. Background
SCN2A is a voltage-gated sodium channel gene that encodes the neuronal sodium channel NaV1.2 and plays a critical role in action potential initiation during early neurodevelopment. The latest study demonstrated that it is loss of function mutations that in SCN2A that lead to autism spectrum disorders (ASD), in contrast to gain of function, which leads to infantile seizures (Ben-Shalom 2018).
In this tutorial, we will handle genetic data for SCN2A mutations identified in latest genomic studies, and then explore the data format to describe genetic mutations using R basic functions. Our tutorial will utilize the summary data from Sanders et al. (2018).
1.2. Aims
What we will do with this dataset,
Understand the dataset from a scientific journal
Apply some functions you have learnt from the Chapter 2 and 3
2. Explore your data
2.1. Unboxing your dataset
Here we obtain the list of mutations in the Supplementary Table 1 from Sanders et al. (2018).
Using the rio package, reading the excel file from the file link into your workspace. If you don't have the rio package in your system, please install as following:
install.packages('rio')Now you can read the file from the website. This will create the d object.
Let's explore the object you just loaded. How would you check the class of the object d?
It shows that the d object is data.frame.
Then, let's overview the data frame. We will use head function to print out first few lines.
When you execute code in a notebook chunk, an output will be visible immediately beneath the input. From this, you can see several rows and columns in the data frame.
Let's look at the first column PatientID and check which class it is.
Cool. Now you can see the TrueRecurrence column. What is the class of the column TrueRecurrence?
To check the class of columns, you don't need to type an individual column. We can overview the summary of the dataset using summary function. Which column has the character class?
2.2. Difference between data frame and matrix
Here we will convert the data frame into a matrix, and compare which part will be different in this. To convert a data frame into a matrix, you can use the command called as.matrix.
Let's overview the matrix object. Can you tell difference with data frame?
2.3. Subset and Sort
Some patients who have the SCN2A mutation (hereafter called "SCN2A patient") often have seizures. So we want to know when the seizure occurs in development.
Let's check the class first.
Why this column contains character? Let's head the first few lines.
It seems that some rows contain samples who do not have seizure or unknown information. It's represented by ".", and also recorded in another column called Seizures.
So we want to subset rows where the seizure phenotype is available.
Let's see how many samples have seizure phenotypes? Then, you can ask when is the earliest days for the representation of seizure phenotype? How can we check this? The fisrt, as seen previously the SeizureOnsetDays column is character so we cannot apply functions for numeric.
So we have to convert this into numeric first.
Hmm. There's an warning for NA introduction. This is because some rows do not have character that we can properly convert from character to numeric. So possible solutions are either you can bear with this in your downstream analyses or 2) convert character into an appropriate form of numeric conversion.
Then, the question is how can we find the rows with NA? We will ask whether the rows contains NA or not using is.na function. This will return boolean as to NA presence.
See we can find some rows with NA. One of them is the 13th row. Let's see how it looks like.
Here you have < (angle bracket) in the character so it won't properly converted to numeric information. Did you find more of these cases?
Our NAs all contains <, which prevent converting a character into a numeric.
We would fix for downstream analyses. For example, we can convert <365 into 365. One function we can try is gsub. This replace your string into a format that you may not get NA. For example,
Let's convert them into numerics.
Did you get warning for this? Now we can ask our initial question. When is the earliest day for having seizure?
3. Exercise
The dataset contains more details for genetic mutations in SCN2A patients. From this information, what can we analyze further?
Here I list up few questions you can examine further.
Finding the position of the genetic mutations within SCN2A. Which information you would use? If you are not familiar with positional information on genetic variants (or mutations), please find the Figure 1 or the slides for Mutation (BSMS205 Session 3-1).
Counting the recurrent mutations at the same protein position (in other words, the same mutations seen across different patients), and examine whether the patients have similar phenotype.
Finding the position where different consequences mutations occur. Please note that "consequences" are loss-of-function (Nonsense, Frameshift) or missense.
Sketch a plot to visualize your analysis.
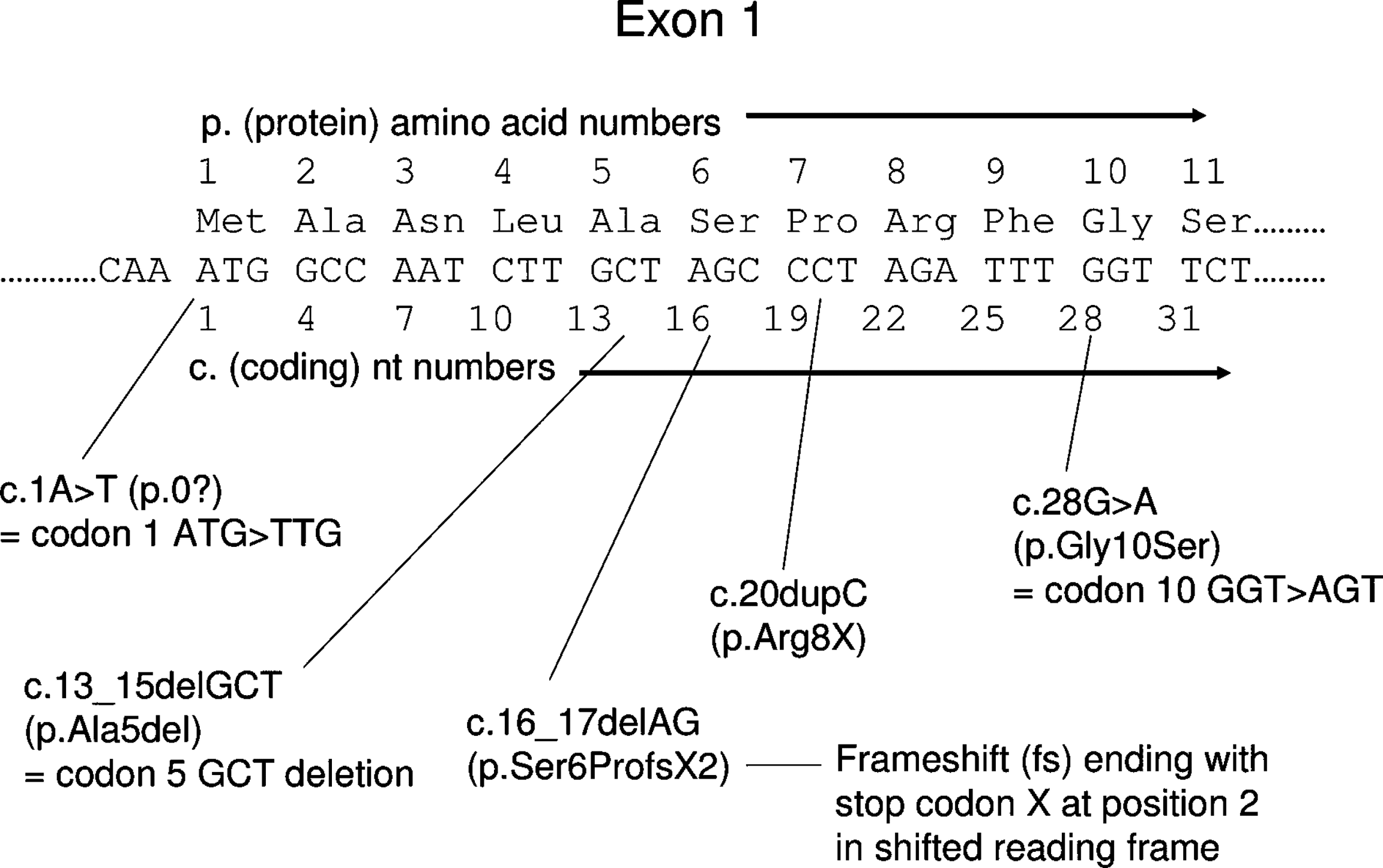
Figure 1. Standard mutation nomenclature (Ogino et al. 2007). According to this guideline, genetic mutations can be represented for coding DNA reference sequences (c. prefix ) and protein-level amino acid sequences (p. prefix).
3.1. For-loops and Vectorization
Here we examine more details of genetic mutations as to their functional consequence and position of SCN2A mutations. In the dataset includes, there are two columns called c.DNA and p.Protein, containing the cDNA or protein position for the genetic mutations.
During these exercises, we will look at the concept of for-loop and vectorization, which you learn from the Chapter 3.4. Let's look at the column p.Protein. It contains protein positions from each patient. What would you check at the first place?
Task 1
Task 2
Task 3
....
Task N
Let's write down your code to explore this column.
If you need more information on SCN2A, please visit the Uniprot description for SCN2A. The Uniprot database contains description for protein domains.
Then, I would remove the characters from the string so we can have only numerics for positions. Here I use gsub function to extract numbers from string. Let's remove non-numeric characters from the string.
In the dataset, we have many rows for protein positions. One way we might try is to set up for-loop to process each row.
Do you think this is an effective approach? As we have done in your assignment, for-loop is not a good choice to process vectors because R can do vectorization for this process with a shorter and clearer code. So this mean you can apply gsub on vector and return your output to another column (could be new assign).
3.2. Counting the recurrent mutations
Recurrent mutations are the ones that the same genetic mutations occur in multiple individuals. Recurrent mutations can be common 1) when the mutation does not affect on natural selection, 2) when the mutation is beneficial, 3) in the hotspot for a disease or strongly associated with trait. However, given we are dealing with the genetic mutations from rare disorders, the mutations in the dataset are supposed to be uniquely present in general population. Otherwise, the recurrent mutations can indicate strong association with the phenotype.
To assess the recurrent mutations, the first thing we can try is to examine whether the same mutations occur in multiple individuals. Since the dataset contains individual patients for each row, we can simply check the frequency using:
or we can check the number of unique variants in the dataset by:
How many unique variants you can find? and which variants are occurred in multiple times?
Then, you can use other columns to check frequency for different groups. Which columns you would use for more grouping?
If you find that, please check the recurrent mutations for each group.
3.3. What is the proportion of diagnosis for SCN2A patient?
SCN2A mutation can have multiple different consequences for disease phenotypes. It can cause ASD but also other neurodevelopmental conditions. In total cases, how many phenotypes occur in SCN2A patients. Then, calculate the proportion of the phenotypes among total cases.
Then, you might be intrigued to whether females and males have different occurrence in each disorder. Let's check it.
Another question you can ask is whether different mutation consequences occur in each phenotype. Let's find out how many mutation consequences are observed in each phenotype.
3.4. Find the position where different consequences of mutations occur
If you checked the recurrent mutations, you might want to find a locus where two or more variants occur. Such loci might indicate functionally important position of the gene and you might find some insight as to a cause of disease.
3.5. Sketch a plot to visualize your analysis
When you examine the dataset, you would draw something to show your output. Though we haven't learnt how to plot data yet, we can have a quick sketch for the dataset. There's no restriction on your suggestion. Please submit your hand-drawing for the plot you would like to show from this dataset.
Last updated